Molecular Techniques in Life Science students and the X-Yeast: Course review in MeMoLS
As the second semester of the Molecular Techniques in Life Science (MTLS) Joint Master Programme is comming to an end it’s time for another course review! During the months of March and April we had a course in Methods in Molecular Life Science, or MeMoLS as we know it. This course mixes a bit of everything, including lectures, wet lab and dry lab; and it is all about techniques commonly used in molecular biology.
The lectures
The MeMoLS course is part of a bigger course (called Methods and Concepts in Molecular Life Science) organized by the Department of Molecular Biosciences at Stockholm University, so we had classes with some other last-year bachelor students from the department. During the twelve lectures we covered several basic techniques used everyday in molecular biology labs: how to study protein interactions, using yeast as a model organism or transcriptomics were some of them. Althought the contents were not new for those of us with a background in biological sciences, many of the lectures are great researchers and enthusiastic professors and were very friendly with us every time we approached them during the breaks to ask about their scientific work.
Wet lab: yeast mutagenesis
The practical part of the course was the best of the MeMoLS course, especially because we got to create our own datasets and perform a whole bioinformatics analysis on them!
We started the practical by doing random mutagenesis on some yeast. Just as the X-men, we were aiming to get X-yeasts with mutations in a X-gene (in our case three genes involved in a common pathway: lysine biosynthesis). For this, we cultured our cells in the lab several days, did random mutagenesis and several analysis to check that our yeast were mutants, and to select our lysine-mutants of interest.
Once we had our mutant strains prepared we extracted their DNA and the lab assistants did a whole genome sequencing (WGS) during the spring break so we had fresh results to analyze when we got back from holidays.
All of this took us around two weeks of lab-afternoon sessions every two days. I had never worked with yeast before, so it was fun to learn how to use a new model system, and I quite liked it!
Dry lab: searching for our mutations
The last part of the course consisted on finding which specific mutations one of our yeast strains had. For this we did a whole bioinformatics pipeline: from checking the quality of the WGS reads, to mapping it against a reference genome and calling the variants. We learned to use many bioinformatic tools and softwares used in a daily-basis by expert bioinformaticians, so it was a perfect training!
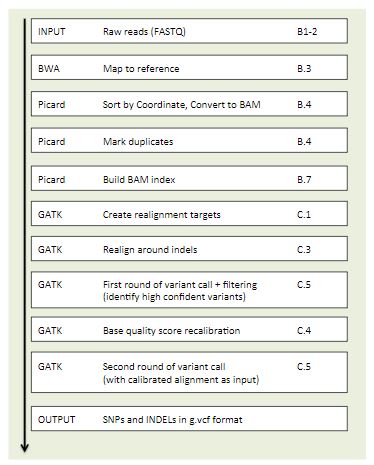
It was a bit challenging to perform and understand all the steps by ourselves, but we had the help of some amazing teaching assistants that guided us thought the process and were very helpful everytime we had a question or a technical problem arose. We also got to use Uppmax, Uppsala’s hight-performance computers, which is something that many bioinformaticians in Sweden use.
The best of the course and my advice for future students
My favourite part of the course was the practical: it was nice to have some wet lab work, and bioinformatics is always fun! I learned so much and enjoyed while doing so, and since we woked in teams of 3 it was the perfect environment for discussions regarding how to run or write the codes and about the results we obtained.
Lastly, my advice for future students is: try to take as much as you can out of the bioinformatics practical. You will learn things totally useful and that you will apply in your future job (if you still do any computation or data analysis after the master’s!).
Please send me all your questions about the MTLS programme, or life in KI/Stockholm in general!
/Inés
email: ines.rivero.garcia@stud.ki.se
LinkedIn: Inés Rivero García

0 comments