Creating a gene predictor: course review in Comparative Genomics
Comparative Genomics has been the last course of the second semester of the Molecular Techniques in Life Science (MTLS) Master Programme. During this course, taught in Stockholm University, we put into practice our coding skills by doing many practical work in the fields of phylogenetics and phylogenomics, network biology and gene prediction amongst others!
How is the course organised? Lots of team work!
This course contains a lot of practical work. Most of what we do during the five weeks that the course spans is to answer computer practicals and write a final project. For this, we are randomly divided into teams, which are kept until the end of the course.
The Comparative Genomics course has three components:
- Lectures and readings.
- Practicals
- Finals project
Lectures
We had a lecture once a week, on Mondays. This served as an introduction to the topics that we would be working with during the week.
In order to prepare for the lecture, we were given in advance several articles, reviews, book chapter(s) and/or web-pages we had to read so that we had some understanding of the field before the lecture. Also, before the start of every lecture we had a small quizz to test what we had learned during the readings
Practicals
During the very first session of the course we were divided into random groups of three people. These groups were kept until the end of the course, and most of the course load was based on solving problems in our teams and writing a report with our answers.
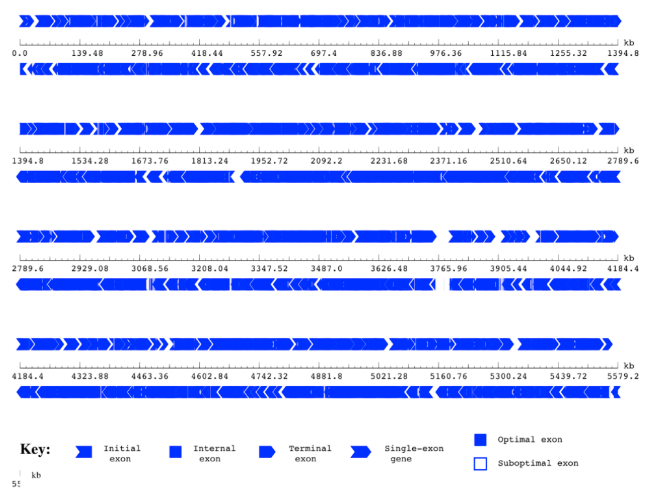
Gene map predicted by using GeneScan
Every week we were introduced to one or two practicals. We had one week to answer all questions in both of them, which included a mixture of theoretical questions, coding problems and tasks for which we had to use databases or specialised software. The topics of the practicals were:
- Basic Genome Analysis: we looked for information about five unidentified genomes and used several different strategies to perform sequence alignments amongst them.
- Gene Prediction: we used two gene predictors, Glimmer and Genescan, to find the predicted genes in our five genomes and analyse gene the gene length distribution and other properties of our genomes.
- Phylogenetic Reconstruction: we used several methods to reconstruct the phylogeny of our five genomes and compared the similarities, differences and statistical support of the different methodologies.
- Phylogenomics: we used groups of homologous sequences in our genomes to reconstruct their phylogeny using a metagene and a consensus tree approach.
- Gene Order Analysis: we studied the conservation of gene order between our assigned genomes.
- Orthology Detection: we differentiated between the orthologs and different types of paralogs in the different genomes.
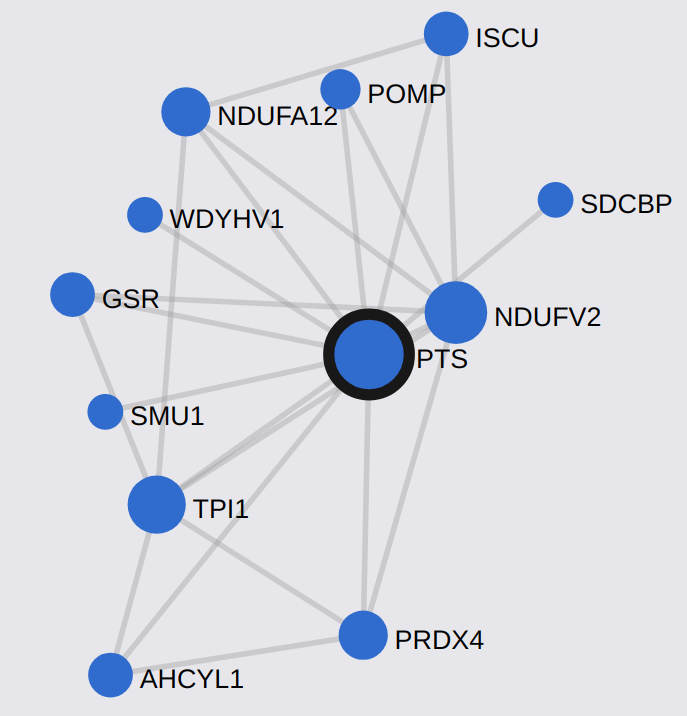
- Interaction Networks: we studied the properties of different biological networks and became familiar with some databases and software used in this field.

We used the database STRING to study protein networks
Final project
To end the course, we did a Final Project in our teams in which we had to use our coding skills to write three small programmes for genome analysis. It was a very intense week of work, since we had a bit more than one week to create the programmes, analyse the data and prepare a project report and presentation.
We all presented our programmes and results to the rest of the class, and it was great to see the different strategies and approaches used by the different groups.
My advice for future students
The Comparative Genomics course is the most coding-intensive course we have had in the MTLS programme so far. This means that, as fun as coding can be, sometimes it’s hard to make the programmes work and we have to learn to deal with frustration. However, this also brings up very interesting discussions to figure out how to make a script work or what is the better approach to tackle a problem.
For me, the best part of the course was that most of it is practical work, which helps to understand and put into context what we read and have learned in previous courses. Also, it is a great opportunity to become a better bioinformatician, and if the group members have the same amount of interest and workload the course is a lot of fun.
Lastly, if you want to take full advantage of the course I’d suggest to do all the practicals as a group instead of dividing the work. It surely slows things down, since one exercise is done at a time, but you will learn a lot from discussing programming with your classmates and writing scripts with them. Also, the readings were useful for me to understand the practicals and to answer them properly, so be sure to read before every lecture!
Any questions about the MTLS programme or life in Stockholm? Drop me an email 🙂
\Inés
email: ines.rivero.garcia@stud.ki.se
LinkedIn: Inés Rivero García

0 comments