New year, new beginnings: starting the Molecular Techniques in Life Science Semester two
New year means new beginnings. For us, the Molecular Techniques in Life Science (MTLS) students, the arrival of the new year has meant that we have started our second semester, which takes place at the Frescati campus of Stockholm University. With all the excitement – and getting lost while searching for lecture rooms – of moving to a new campus, we have also been very busy while taking our bioinformatics course. This course has been the most demanding so far, but it has also taught extra computer skills.
Semester 2 of the MTLS Programme: Stockholm University
The second semester of the MTLS programme takes place in the Department of Biophysics and Biochemistry of Stockholm University (SU), and so we have courses that focus on bioinformatics, biochemistry and biological membranes, as well as some computational genomics and molecular biology techniques.

Views from the library at the Frescati campus
How the semester is organized at SU is quite different to how KI works. Our courses at SU last only about 4 or 5 weeks, which means that during that time we are fully dedicated to a single course, and the final exam comes right at the end of it. Some people like more this system, other people like it less… It has the advantage that it makes you work every day to avoid losing track of the course, but the high pace makes it difficult to catch up if you are lost with a part of the course.
Bioinformatics: the first challenge of 2019
The very first course we had at SU was bioinformatics, a 7 ECTS course focused on basic bioinformatic techniques (sequence comparison, phylogenetic studies and machine learning amongst others) as well as studying how can we take advantage of computers to predict the tridimensional structure of proteins and RNAs. The course lasted four weeks in total, the first two of them were dedicated to these basic bioinformatic techniques I mentioned, and the second half was dedicated to structural bioinformatics. We had two partial exams, one for each block.
The course is organized as a “flipped-classroom”, which means that before each lecture we had some readings to do (chapters of books, papers…) to prepare ourselves for the lecture. Also, we had some short “pre-lecture quizzes” which aimed to review the knowledge gained during the readings and that we had to submit before every day’s lecture. Apart from these preparations and the lectures themselves, we also had computer labs and a project(the two best part of the course in my opinion); I’ll talk about these later on this post.
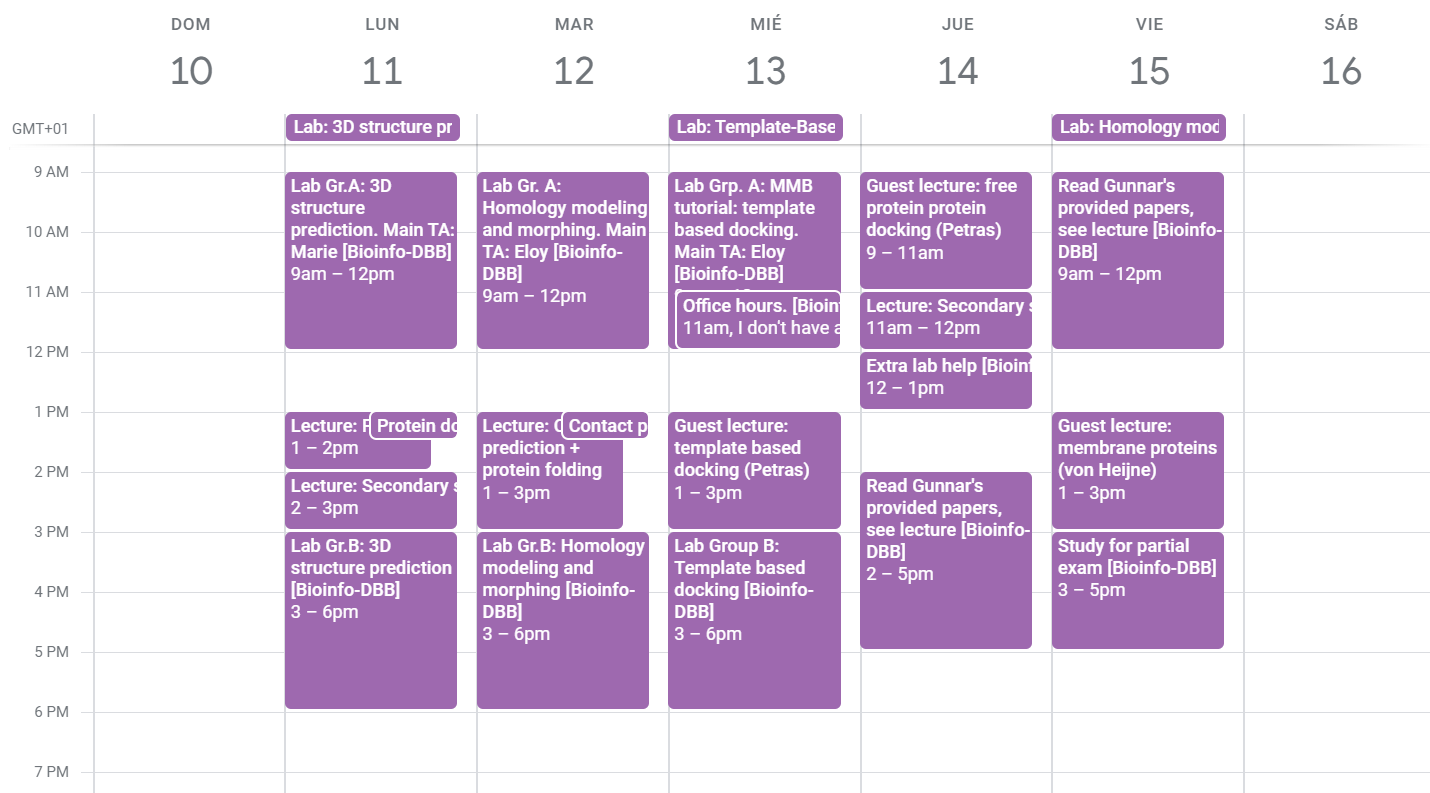
The course schedule from the 10-16 of February. There were two lab groups, so we only needed to attend one lab session.
Keeping up with the readings and lectures
Since we read before every lecture, the lecture’s purpose was more to give a summary and overview of the topic and answer our questions that to teach from scratch. It was an advantage to have read the topic before the lecture because we had the chance to ask all our questions and to understand it better. Also, all the lectures were recorded in video and uploaded by the professors, and that came to be very useful when studying for the exam: it was always possible to re-watch that parts that you didn’t understand or to take better notes of them.
From my experience, the hardest part of this course was keeping up with the readings. I really liked the topics we covered throughout the course and I learned many new and interesting things (which also broadened my interests for future projects!), but it was very time-consuming to go though every assigned reading in enough depth to understand it. That being said, I enjoyed the readings very much and felt like I was very positively investing my studying time.
Appart from the normal lectures we had some guest speakers who are top researchers in their fields: Carsten Daub taught about transcriptomics, Gunnar Von Heijne about prediction of membrane protein topology and Petras Kundrotas about protein docking.
Computer labs: from BLAST to modelling protein structure
Each one of the lectures was followed by a computer lab in which we put into practice our bioinformatics skills, and we also got to practice some bash programming. The computer labs were when I learned the most and when I understood many thinks that I felt quite unsure about during the lectures. We did some computer work to create a gene phylogeny, cluster samples by their mean gene expression, model the 3D structure of RNAs and proteins and docking a protein with its ligands.
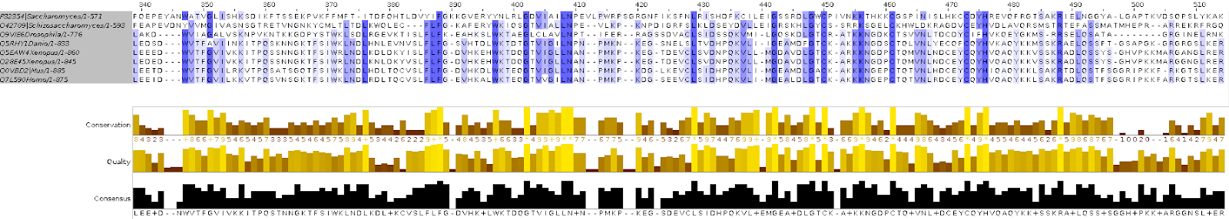
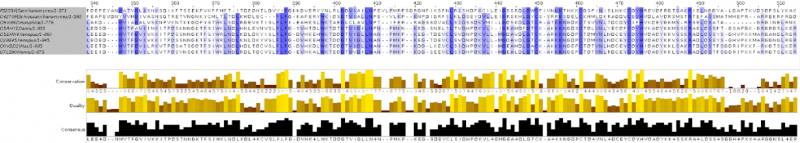
In one of the computer labs we had to build a phylogeny from protein sequences.
For the computer labs we had the help of four PhD students as teaching assistants (TAs). They were such a big help, and very patient because some days we had an immense amount of questions.
Secret sequence: all-in-one project
To finish the course we had to submit a project which completely summarized all that we had learned in the course, and which was fun! We were given a “secret” protein sequence, and by secret I mean that we had no identifiers or any kind on information about it other than the sequence itself, and we had to use all the bioinformatics tools that we had learned about the course to obtain information about this sequence. For example, we had to identify the name of the protein, which organism does it belong to, create a phylogeny for it, identify its structural domains and create a 3D structure model based on a similar homolog. What I most liked about this project is that it felt like doing real research and that there were unlimited options to explore: we had some compulsory points to cover, but then you could explore those tools or questions that were more interesting.
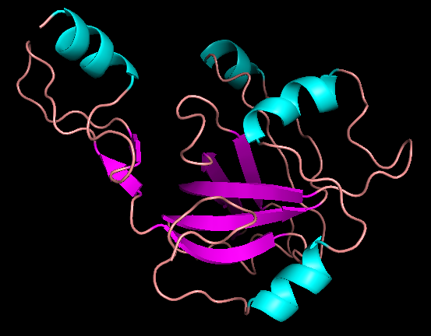
The 3D model I built for my secret sequence.
Overall, I really enjoyed what I learned during this course, despite its very high intensity and the amount of work we had to do. So far it has been the course in which I feel I had learned the most during the MTLS Programme, so the effort has been totally worthwhile!
Feel free to contact me for any questions 🙂
/Inés
Email: ines.rivero.garcia@stud.ki.se
LinkedIn: Ines Rivero Garcia

0 comments