Course review in Applied Gene Technology and Large-Scale Data Analysis
Have you ever felt amazed by the new methods developed in a molecular biology paper? Althoug I have no experience as a wet or dry lab method developer, I am always inspired by the researchers who develop new cheaper, smarter or more efficient technologies. This is probably one of the reasons why I enjoyed so much the course in Applied Gene Technology and Large-Scale Data Analysis.
My personal view on the course
From my perspective, it was very easy to get engaged in this course since the topics were interesting and it has a good balance of work that demands the students to work everyday a bit on it without feeling very overwhelmed by the workload. I specially liked the critical point of view that was continously applied during the GeneTech block, and the usefulness of the data analysis lectures.
What is the couse about?
As it can be guessed from the title, the course has two parts:
- Applied Gene Technology: we covered topics on molecular genetics, measurement of gene expression and sequencing.
- Large-Scale Data Analysis: includes topics in genome assembly, gene mapping, and bulk RNA-Seq, scRNA-Seq and spatial transcriptomics data analysis.
The lectures
In the GeneTech block, we had six lectures that went from the molecular toolbox to the newest sequencing technologies in a chronological order. Most of the concepts were already present in other courses of the master’s, but I liked that we had to think about them more critically and had to answer questions that I had never thought of, even though I had understood the technique before! The lectures were not only focused on the theoretical background of the techniques, but rather on critically thinking about them to identify their pros and cons and find the applications for which they are more suitable.
The bioinformatics part was a bit more theoretical, since it was complemented by computer labs in which we put all our programming skills into practice. The first lectures (genome assembly and read mapping) had already been covered in the Bioinformatics course, but in this occasion we read about more different algorithms, and we got a broader view of the state of the field. On the other hand, the three lectures in data analysis were eather new for us, and I specially liked them because they were very easy to relate to the biological part of the course, and some of us had already encountered the problems discussed in class and got the chance to understand better the log behind some pipelines and algorithms.

The Applied Gene Technology and Large-Scale Data Analysis is a great example of how the MTLS programme combines biology with programming
Critically reading methods articles: bibliographical project
Half of the GeneTech block consisted on a project in which we had to read a series of scientific articles, present the methods developed by the authors and critically asses their strenghts and weaknesses. We did this in groups of four people, and we covered four topics: hybridization technologies, long-read sequencing, single cell technologies and spatial transcriptomics.
We had many chances to discuss about our specific project with both our teammates and professor, Patrick Ståhl, so that we felt quite confident about the topic we had to present before the presentation day arrived. Even though we were supposed to focus on one of these four topics, we all had to read about the four topics and submit questions for our fellow classmates to answer. All in all, it was a great chance to read many scientific papers and discuss about them. Also, some of them were super interesting!
Hands-on data analysis: computer labs
The practical block of the data analysis half consisted in, yes, you guessed it, computer labs! This time we used the language R, which was a bit confusing at the beginning since in all of the other courses of the master’s we have used Python.
We had three computer practicals in which we analyzed data from bulk RNA-Seq, scRNA-Seq and spatial transcriptomics experiments. Moreover, one of the labs included a extra part in which we had to analyze the results of some GWAS experiments to understand how to get from the statistical data to the biological meaning of it.
I learned a lot while doing these practicals with my classmate Miren, and it was great practice since bioinformaticians analyze thise type of data on their daily basis.
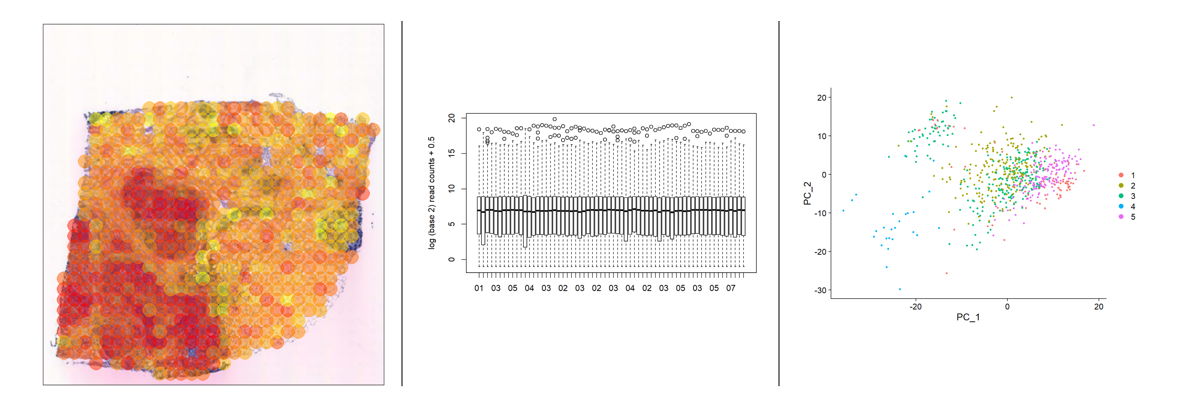
Data Analysis from Spatial Trasncriptmics (left), bulk RNA-Seq (center) and scRNA-Seq (right). Credits: Miren Urrutia Iturritza and myself!
I hope this course review was useful for those of you interested in the Molecular Techniques in Life Science Joint Master Programme. Please email me all your questions about the programme or life in Stockholm.
\Inés
email: ines.rivero.garcia@stud.ki.se
LinkedIn:Inés Rivero García

0 comments